Structural Analysis of Viruses by HDX MS
Background & Overview
Structural analysis of viruses is a complex task. The details of viral structure, especially enveloped viruses, whose structural proteins and viral assemblies tend to have multiple conformational states and undergo dramatic structural changes make them difficult to study by most structural methods. Many viruses also frequently include highly dynamic regions of critical functional importance, as well as large surface glycoproteins, which have also been shown to increase the challenge of studying their structural biology. Hydrogen-deuterium exchange coupled with mass spectrometry (HDX MS) can provide a wealth of information about many different viral proteins, glycoproteins, and complexes. BOC Sciences has developed methods to study the rich structural dynamics of viral systems using HDX MS.
Our Services
We use Ultra High-Pressure Liquid Chromatography (UPLC), which provides higher chromatographic resolution and greatly increases the complexity of the samples that can be analyzed. Not only that, but the use of modern mass spectrometers and HDX MS data processing software has expanded the limits on sample complexity, greatly facilitating the analysis of experimental data while reducing detection limits. Our virus structure analysis services include but are not limited to:
- Conformational purity assessment and biosimilar comparison
- Structural studies of membrane proteins
- Study of structural changes in viral capsids, comparing capsids in different conformational states, helping to pinpoint regions undergoing significant local reorganization
- Local conformational kinetic measurements and studies on the assembly and capsid maturation mechanisms of various phages, including human immunodeficiency virus, sarcoma virus, adenovirus, hepatitis B, brome mosaic virus, rhinovirus, etc.
- Bimodal spectral detection, reflecting the different conformations that a given region of a protein may exist in a sample
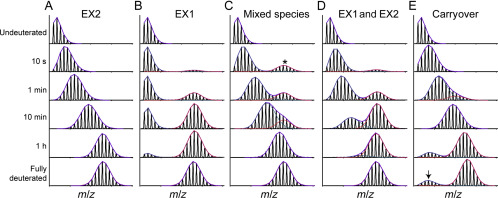 Fig.1 Examples of different exchange behaviors during HDX-MS.[1]
Fig.1 Examples of different exchange behaviors during HDX-MS.[1]
- Informative analysis of intrinsically disordered proteins in solution, such as elucidating residual structures present in nonstructural proteins of HIV Nef and Dengue virus
- Detection and analysis of viral glycoproteins, including hepatitis C E2 glycoprotein, influenza hemagglutinin, HIV envelope glycoprotein, and Ebola GP1/GP2, etc.
Our Methods
- First, the exchange reaction is initiated by diluting the protein stock solution into deuterated buffer, and an appropriate deuteration time is selected.
- Then, after a specific time of deuteration, the amide exchange is slowed down (or "quenched") by reducing the temperature to ~0 °C and pH to 2.5, ensuring that the protein can remain intact for the overall deuterium check.
- Next, quench and digest conditions are optimized to provide the highest possible number of observable peptides to ensure maximum sequence coverage. For example, guanidine urea is added to the quench solution to help unfold the protein. Certain nonionic detergents can also be added to the buffer at low concentrations compatible with LC-MS to improve solubility or stability during deuterium exchange.
- Deuterium incorporation was measured by mass spectrometry after the samples were digested. Quench conditions were maintained throughout the analysis to avoid further back exchange. Using electrospray ionization (ESI) in combination with reversed-phase UPLC provides greater sequence coverage and more complete structural analysis.
- Electron transfer dissociation is used instead of MS/MS tools to identify glycopeptides. Samples were re-neutralized after digestion, treated with endoglycosidase to remove glycans, and collision-induced dissociation fragmentation data were obtained for deglycosylated peptides.
- Finally, we used HDX data analysis software to determine deuterium absorption for a given peptide, including hundreds to thousands of spectra. Techniques using overlapping peptides have also been developed to obtain higher resolution exchange information.
If you are interested in our services, please contact us immediately, then fill in the complete inquiry form, and we will reply to you as soon as possible.
Reference
- Guttman M, Lee K K. Isotope labeling of biomolecules: structural analysis of viruses by HDX-MS[M]. Methods in enzymology. 2016, 566: 405-426.


 Fig.1 Examples of different exchange behaviors during HDX-MS.[1]
Fig.1 Examples of different exchange behaviors during HDX-MS.[1]
